G3-1 バイオマーカーおよび治療法開発を加速するデータ駆動型モデリング
Functional analysis of B and T cell receptor sequence data, RNA- and DNA-binding protein modeling, multiple sequence alignment
[1] 支援担当者
| 所属 | ①大阪大学 微生物病研究所遺伝情報実験センターゲノム情報解析分野 | |
|---|---|---|
| 氏名 | ①Daron Standley, 川口 美恵、都 雅子 | |
| AMED 事業 |
ユニット/領域名 課題名 |
インシリコユニット バイオマーカーおよび治療法開発を加速するデータ駆動型モデリング |
| 代表機関 代表者 |
大阪大学 Daron M. Standley |
|
| 支援技術のキーワード | antibody, antigen, sequencing, structure, RNA | |
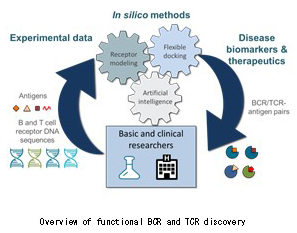
[2] 支援技術の概要
We are developing various bioinformatics techniques to support the discovery of immune-based biomarkers and therapeutics. To this end, we can perform high-throughput receptor modeling, flexible docking and functional classification of results using AI.
[3] 支援技術の利用例
- High-throughput BCR and TCR modeling
- Structural clustering of BCR/TCR models
- Prediction of nucleotide binding sites
- Flexible nucleotide-protein docking
- Multiple sequence & structure alignment
[4] 支援担当者の研究概要
The research in our laboratory is concentrated in three main areas: (1) Analysis of B cell receptors (BCRs) and T cell receptors (TCRs) from large-scale sequence data; (2) modeling protein-nucleotide interactions, especially those that mediate immune responses; (3) multiple sequence and structural alignment.
Analysis of BCRs and TCRs from large-scale sequence data. BCRs and TCRs undergo molecular evolution within the lifetime of the host organism in response to antigen exposure. They also represent the fastest-growing class of therapeutic molecules in a wide range of disease areas. We are thus focusing on developing methods that can harness the Big Data from BCR and TCR sequencing studies to develop biomarkers and therapeutics. We make use of recent technology developed in our lab for modeling BCRs and TCRs to atomic resolution in a short amount of time (e.g., seconds).
Modeling protein-nucleotide interactions that mediate immune responses. Immune cells regulate the strength and duration of responses to external stimuli both at transcriptional and post-transcriptional levels. Of these, the post-transcriptional regulatory pathways that act via RNA-binding proteins (RBPs) have only recently been appreciated. In order to identify novel RBPs and their modes of action, we have developed a class of tools that can flexibly dock single-stranded RNA and DNA molecules to proteins using only sequence information (for both nucleotide and protein as input).
Multiple sequence and structural alignment. We develop the MAFFT multiple sequence and structure alignment package, which is one of the most popular and versatile alignment methods available.